Difference between revisions of "Publications/xu.17.icip.inc"
From LRDE
Yongchao Xu (talk | contribs) |
Yongchao Xu (talk | contribs) (→Method) |
||
| (54 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | == Method and datasets == |
||
| + | |||
| + | === Method === |
||
| + | |||
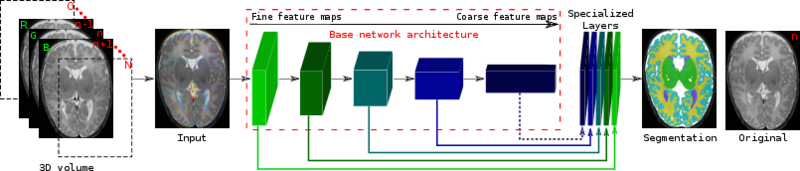
| + | Architecture of the proposed network. We fine tune it and combine linearly fine to coarse feature maps of the [http://www.robots.ox.ac.uk/~vgg/research/very_deep/ pre-trained VGG network]. The coarsest feature maps are discarded for the adult images. |
||
| + | |||
| + | [[File:Xu.17.icip-pepeline.png|800 px]] |
||
| + | |||
| + | === Datasets === |
||
| + | * Dataset of the MICCAI challenge of Neonatal Brain Segmentation 2012 ([http://neobrains12.isi.uu.nl/ NeoBrainS12]) |
||
| + | ** Axial images acquired at 40 weeks: 2 training images + 5 test images |
||
| + | ** Coronal images acquired at 30 weeks: 2 training images + 5 test images |
||
| + | ** Coronal images acquired at 40 weeks: 5 test images |
||
| + | * Dataset of the MICCAI challenge of MR Brain Image Segmentation ([http://mrbrains13.isi.uu.nl/ MRBrainS13]) |
||
| + | ** Axial images acquired at 70 years: 5 training images + 15 test images |
||
| + | |||
== Materials == |
== Materials == |
||
| + | |||
| + | === Trained models === |
||
| + | The trained models and corresponding files for training for the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following: |
||
| + | * Training on Axial images at 40 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_axial40_model.zip archive] |
||
| + | * Training on coronal images at 30 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_coronal30_model.zip archive] |
||
| + | * Training on previous training sets for coronal images at 40 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_coronal40_model.zip archive] |
||
| + | * Training on MRBrainS13 dataset is available in this [https://www.lrde.epita.fr/~xu/material/mrbrains13_model.zip archive] |
||
| + | |||
| + | === Segmentation results === |
||
| + | The pre-computed segmentation results of the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following: |
||
| + | * Results on Axial images at 40 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_axial40_seg_results.zip archive] |
||
| + | * Results on coronal images at 30 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_coronal30_seg_results.zip archive] |
||
| + | * Results on coronal images at 40 weeks in NeoBrainS12 dataset are available in this [https://www.lrde.epita.fr/~xu/material/neobrains12_coronal40_seg_results.zip archive] |
||
| + | * Results on MRBrainS13 dataset are available in this [https://www.lrde.epita.fr/~xu/material/mrbrains_seg_results.zip archive] |
||
== Illustrations == |
== Illustrations == |
||
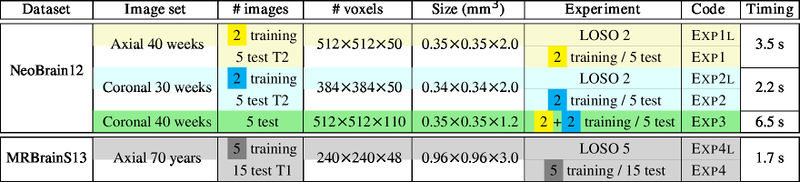
| + | === Experiments === |
||
| + | Leave-One-Subject-Out (LOSO) cross-validation on N images + normal training/test experiments. Note that only '''one''' training image is used for LOSO 2. |
||
| + | [[File:xu.17.icip-experiments.jpg|800 px]] |
||
=== LOSO experiments === |
=== LOSO experiments === |
||
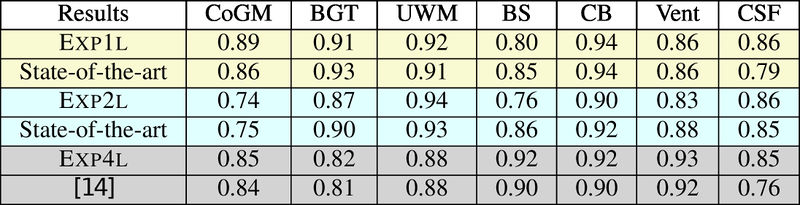
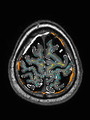
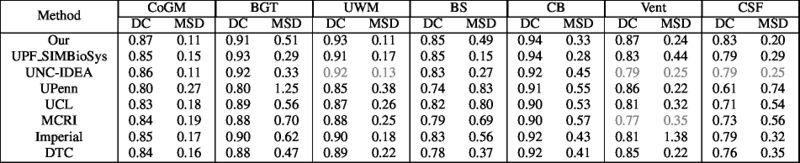
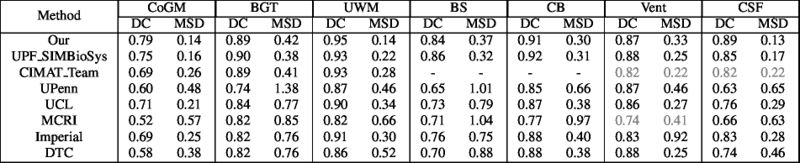
| + | Quantitative results of LOSO experiments in terms of Dice coefficient as compared to the state-of-the-art results. The last one is from [http://ieeexplore.ieee.org/document/7444155/ P. Moeskops et al.] on the 15 test images in MRBrainS13 dataset. |
||
| + | |||
| + | [[File:xu.17.icip-losoresults.jpg | 800 px]] |
||
| + | |||
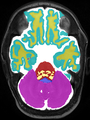
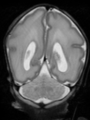
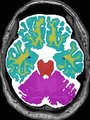
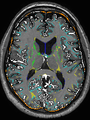
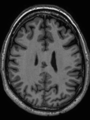
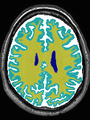
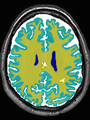
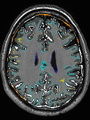
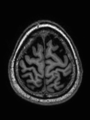
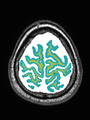
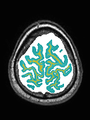
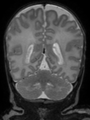
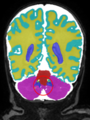
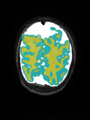
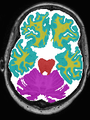
| + | * Some results on axial images at 40 weeks in NeoBrainS12 dataset |
||
| + | |||
<gallery> |
<gallery> |
||
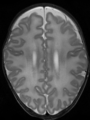
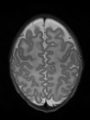
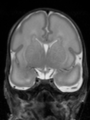
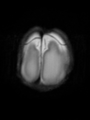
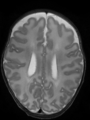
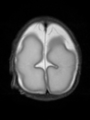
File:xu.17.icip-axialTrain407Input.png |
File:xu.17.icip-axialTrain407Input.png |
||
| Line 9: | Line 48: | ||
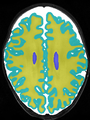
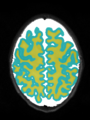
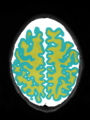
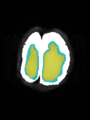
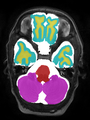
File:xu.17.icip-axialTrain407Gt.png |
File:xu.17.icip-axialTrain407Gt.png |
||
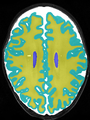
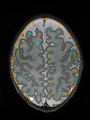
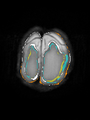
File:xu.17.icip-axialTrain407Diff.png |
File:xu.17.icip-axialTrain407Diff.png |
||
| + | </gallery> |
||
| − | |||
| + | <gallery> |
||
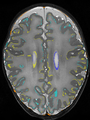
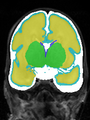
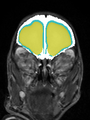
File:xu.17.icip-axialTrain420Input.png |
File:xu.17.icip-axialTrain420Input.png |
||
File:xu.17.icip-axialTrain420Seg.png |
File:xu.17.icip-axialTrain420Seg.png |
||
File:xu.17.icip-axialTrain420Gt.png |
File:xu.17.icip-axialTrain420Gt.png |
||
File:xu.17.icip-axialTrain420Diff.png |
File:xu.17.icip-axialTrain420Diff.png |
||
| + | </gallery> |
||
| − | |||
| + | <gallery> |
||
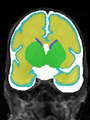
File:xu.17.icip-axialTrain430Input.png |
File:xu.17.icip-axialTrain430Input.png |
||
File:xu.17.icip-axialTrain430Seg.png |
File:xu.17.icip-axialTrain430Seg.png |
||
File:xu.17.icip-axialTrain430Gt.png |
File:xu.17.icip-axialTrain430Gt.png |
||
File:xu.17.icip-axialTrain430Diff.png |
File:xu.17.icip-axialTrain430Diff.png |
||
| + | </gallery> |
||
| − | |||
| + | <gallery> |
||
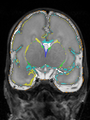
File:xu.17.icip-axialTrain438Input.png | Input |
File:xu.17.icip-axialTrain438Input.png | Input |
||
File:xu.17.icip-axialTrain438Seg.png | Segmentation |
File:xu.17.icip-axialTrain438Seg.png | Segmentation |
||
File:xu.17.icip-axialTrain438Gt.png | Manual segmentation |
File:xu.17.icip-axialTrain438Gt.png | Manual segmentation |
||
File:xu.17.icip-axialTrain438Diff.png | Difference |
File:xu.17.icip-axialTrain438Diff.png | Difference |
||
| + | </gallery> |
||
| + | |||
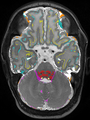
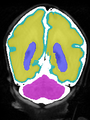
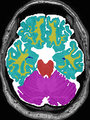
| + | * Some results on coronal images at 30 weeks in NeoBrainS12 dataset |
||
| + | <gallery> |
||
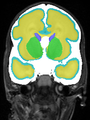
| + | File:xu.17.icip-coronalTrain108Input.png |
||
| + | File:xu.17.icip-coronalTrain108Seg.png |
||
| + | File:xu.17.icip-coronalTrain108Gt.png |
||
| + | File:xu.17.icip-coronalTrain108Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:xu.17.icip-coronalTrain118Input.png |
||
| + | File:xu.17.icip-coronalTrain118Seg.png |
||
| + | File:xu.17.icip-coronalTrain118Gt.png |
||
| + | File:xu.17.icip-coronalTrain118Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:xu.17.icip-coronalTrain131Input.png |
||
| + | File:xu.17.icip-coronalTrain131Seg.png |
||
| + | File:xu.17.icip-coronalTrain131Gt.png |
||
| + | File:xu.17.icip-coronalTrain131Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:xu.17.icip-coronalTrain143Input.png | Input |
||
| + | File:xu.17.icip-coronalTrain143Seg.png | Segmentation |
||
| + | File:xu.17.icip-coronalTrain143Gt.png | Manual segmentation |
||
| + | File:xu.17.icip-coronalTrain143Diff.png | Difference |
||
| + | </gallery> |
||
| + | |||
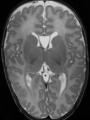
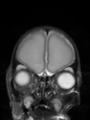
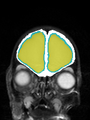
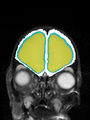
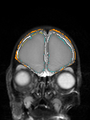
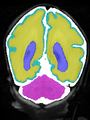
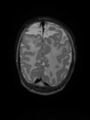
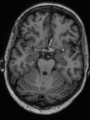
| + | * Some results on axial images of aging adults at 70 ages in MRBrainS13 dataset |
||
| + | <gallery> |
||
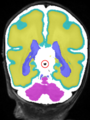
| + | File:Xu.17.icip-mrbrainsTrain513Input.png |
||
| + | File:Xu.17.icip-mrbrainsTrain513Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTrain513Gt.png |
||
| + | File:Xu.17.icip-mrbrainsTrain513Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-mrbrainsTrain522Input.png |
||
| + | File:Xu.17.icip-mrbrainsTrain522Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTrain522Gt.png |
||
| + | File:Xu.17.icip-mrbrainsTrain522Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-mrbrainsTrain527Input.png |
||
| + | File:Xu.17.icip-mrbrainsTrain527Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTrain527Gt.png |
||
| + | File:Xu.17.icip-mrbrainsTrain527Diff.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-mrbrainsTrain538Input.png | Input |
||
| + | File:Xu.17.icip-mrbrainsTrain538Seg.png | Segmentation |
||
| + | File:Xu.17.icip-mrbrainsTrain538Gt.png | Manual segmentation |
||
| + | File:Xu.17.icip-mrbrainsTrain538Diff.png | Difference |
||
| + | |||
</gallery> |
</gallery> |
||
=== Neonatal brain MR image segmentation === |
=== Neonatal brain MR image segmentation === |
||
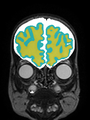
| + | * Results on axial images at 40 weeks in NeoBrainS12 dataset. More details can be found [http://neobrains12.isi.uu.nl/mainResults_Set1.php Here] |
||
| + | [[File:Xu.17.icip-axial40results.jpg | 800 px]] |
||
| + | |||
| + | Some result images |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-axialTest408Input.png |
||
| + | File:Xu.17.icip-axialTest418Input.png |
||
| + | File:Xu.17.icip-axialTest430Input.png |
||
| + | File:Xu.17.icip-axialTest440Input.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-axialTest408Seg.png |
||
| + | File:Xu.17.icip-axialTest418Seg.png |
||
| + | File:Xu.17.icip-axialTest430Seg.png |
||
| + | File:Xu.17.icip-axialTest440Seg.png |
||
| + | </gallery> |
||
| + | |||
| + | |||
| + | * Results on coronal images at 30 weeks in NeoBrainS12 dataset. More details can be found [http://neobrains12.isi.uu.nl/mainResults_Set2.php Here] |
||
| + | [[File:Xu.17.icip-coronal30results.jpg | 800 px]] |
||
| + | |||
| + | Some result images (some small errors inside the red circle) |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-coronal30Test509Input.png |
||
| + | File:Xu.17.icip-coronal30Test517Input.png |
||
| + | File:Xu.17.icip-coronal30Test529Input.png |
||
| + | File:Xu.17.icip-coronal30Test540Input.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-coronal30Test509Seg.png |
||
| + | File:Xu.17.icip-coronal30Test517Seg.png |
||
| + | File:Xu.17.icip-coronal30Test529Seg_circles.png |
||
| + | File:Xu.17.icip-coronal30Test540Seg.png |
||
| + | </gallery> |
||
| + | |||
| + | |||
| + | * Results on coronal images at 40 weeks in NeoBrainS12 dataset. More details can be found [http://neobrains12.isi.uu.nl/mainResults_Set3.php Here] |
||
| + | [[File:Xu.17.icip-coronal40results.jpg | 800 px]] |
||
| + | |||
| + | Some result images (some small errors inside red circles) |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-coronal40Test5021Input.png |
||
| + | File:Xu.17.icip-coronal40Test5054Input.png |
||
| + | File:Xu.17.icip-coronal40Test5070Input.png |
||
| + | File:Xu.17.icip-coronal40Test5094Input.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-coronal40Test5021Seg.png |
||
| + | File:Xu.17.icip-coronal40Test5054Seg_circles.png |
||
| + | File:Xu.17.icip-coronal40Test5070Seg_circles.png |
||
| + | File:Xu.17.icip-coronal40Test5094Seg.png |
||
| + | </gallery> |
||
=== Adult brain MR image segmentation === |
=== Adult brain MR image segmentation === |
||
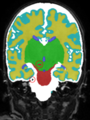
| + | * Results on aging adult images at 70 years in MRBrainS13 dataset. Only top 10 methods among 38 submitted ones are shown. More results and details can be found [http://mrbrains13.isi.uu.nl/results.php Here] |
||
| + | [[File:Xu.17.icip-adult70results.jpg | 800 px]] |
||
| + | |||
| + | Some result images |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-mrbrainsTest514Input.png |
||
| + | File:Xu.17.icip-mrbrainsTest521Input.png |
||
| + | File:Xu.17.icip-mrbrainsTest527Input.png |
||
| + | File:Xu.17.icip-mrbrainsTest537Input.png |
||
| + | </gallery> |
||
| + | <gallery> |
||
| + | File:Xu.17.icip-mrbrainsTest514Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTest521Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTest527Seg.png |
||
| + | File:Xu.17.icip-mrbrainsTest537Seg.png |
||
| + | </gallery> |
||
Latest revision as of 23:04, 6 February 2017
Method and datasets
Method
Architecture of the proposed network. We fine tune it and combine linearly fine to coarse feature maps of the pre-trained VGG network. The coarsest feature maps are discarded for the adult images.
Datasets
- Dataset of the MICCAI challenge of Neonatal Brain Segmentation 2012 (NeoBrainS12)
- Axial images acquired at 40 weeks: 2 training images + 5 test images
- Coronal images acquired at 30 weeks: 2 training images + 5 test images
- Coronal images acquired at 40 weeks: 5 test images
- Dataset of the MICCAI challenge of MR Brain Image Segmentation (MRBrainS13)
- Axial images acquired at 70 years: 5 training images + 15 test images
Materials
Trained models
The trained models and corresponding files for training for the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following:
- Training on Axial images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Training on coronal images at 30 weeks in NeoBrainS12 dataset are available in this archive
- Training on previous training sets for coronal images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Training on MRBrainS13 dataset is available in this archive
Segmentation results
The pre-computed segmentation results of the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following:
- Results on Axial images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Results on coronal images at 30 weeks in NeoBrainS12 dataset are available in this archive
- Results on coronal images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Results on MRBrainS13 dataset are available in this archive
Illustrations
Experiments
Leave-One-Subject-Out (LOSO) cross-validation on N images + normal training/test experiments. Note that only one training image is used for LOSO 2.
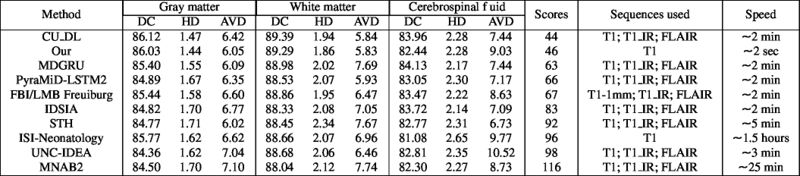
LOSO experiments
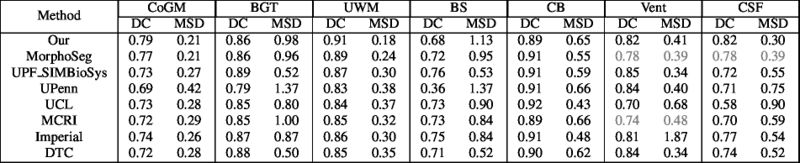
Quantitative results of LOSO experiments in terms of Dice coefficient as compared to the state-of-the-art results. The last one is from P. Moeskops et al. on the 15 test images in MRBrainS13 dataset.
- Some results on axial images at 40 weeks in NeoBrainS12 dataset
- Some results on coronal images at 30 weeks in NeoBrainS12 dataset
- Some results on axial images of aging adults at 70 ages in MRBrainS13 dataset
Neonatal brain MR image segmentation
- Results on axial images at 40 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images
- Results on coronal images at 30 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images (some small errors inside the red circle)
- Results on coronal images at 40 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images (some small errors inside red circles)
Adult brain MR image segmentation
- Results on aging adult images at 70 years in MRBrainS13 dataset. Only top 10 methods among 38 submitted ones are shown. More results and details can be found Here
Some result images