From Neonatal to Adult Brain MR Image Segmentation in a Few Seconds Using 3D-Like Fully Convolutional Network and Transfer Learning
From LRDE
- Authors
- Yongchao Xu, Thierry Géraud, Isabelle Bloch
- Where
- Proceedings of the 23rd IEEE International Conference on Image Processing (ICIP)
- Place
- Beijing, China
- Type
- inproceedings
- Projects
- Olena
- Keywords
- Image
- Date
- 2017-06-12
Abstract
Brain magnetic resonance imaging (MRI) is widely used to assess brain developments in neonates and to diagnose a wide range of neurological diseases in adults. Such studies are usually based on quantitative analysis of different brain tissues, so it is essential to be able to classify them accurately. In this paper, we propose a fast automatic method that segments 3D brain MR images into different tissues using fully convolutional network (FCN) and transfer learning. As compared to existing deep learning-based approaches that rely either on 2D patches or on fully 3D FCN, our method is way much faster: it only takes a few seconds, and only a single modality (T1 or T2) is required. In order to take the 3D information into account, all 3 successive 2D slices are stacked to form a set of 2D color images, which serve as input for the FCN pre-trained on ImageNet for natural image classification. To the best of our knowledge, this is the first method that applies transfer learning to segment both neonatal and adult brain 3D MR images. Our experiments on two public datasets show that our method achieves state-of-the-art results.
Documents
Method and datasets
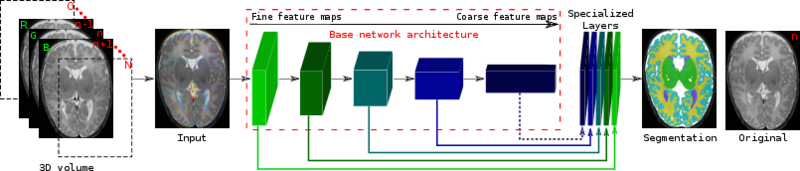
Method
Architecture of the proposed network. We fine tune it and combine linearly fine to coarse feature maps of the pre-trained VGG network. The coarsest feature maps are discarded for the adult images.
Datasets
- Dataset of the MICCAI challenge of Neonatal Brain Segmentation 2012 (NeoBrainS12)
- Axial images acquired at 40 weeks: 2 training images + 5 test images
- Coronal images acquired at 30 weeks: 2 training images + 5 test images
- Coronal images acquired at 40 weeks: 5 test images
- Dataset of the MICCAI challenge of MR Brain Image Segmentation (MRBrainS13)
- Axial images acquired at 70 years: 5 training images + 15 test images
Materials
Trained models
The trained models and corresponding files for training for the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following:
- Training on Axial images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Training on coronal images at 30 weeks in NeoBrainS12 dataset are available in this archive
- Training on previous training sets for coronal images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Training on MRBrainS13 dataset is available in this archive
Segmentation results
The pre-computed segmentation results of the proposed method on NeoBrainS12 and MRBrainS13 datasets are available in the following:
- Results on Axial images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Results on coronal images at 30 weeks in NeoBrainS12 dataset are available in this archive
- Results on coronal images at 40 weeks in NeoBrainS12 dataset are available in this archive
- Results on MRBrainS13 dataset are available in this archive
Illustrations
Experiments
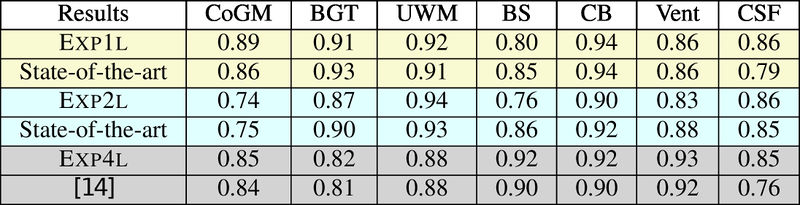
Leave-One-Subject-Out (LOSO) cross-validation on N images + normal training/test experiments. Note that only one training image is used for LOSO 2.
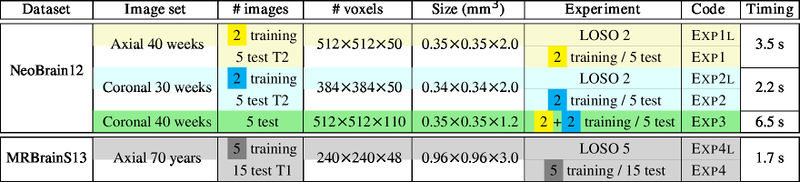
LOSO experiments
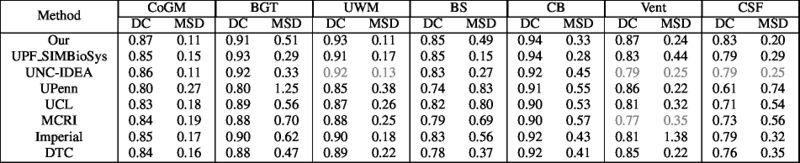
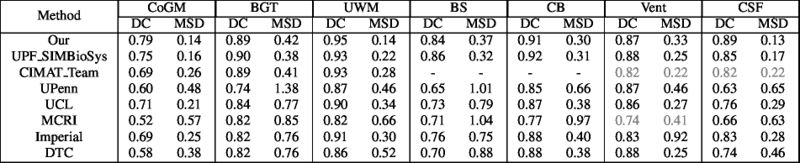
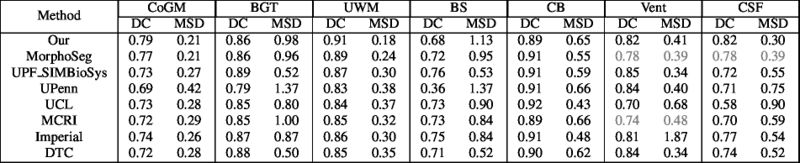
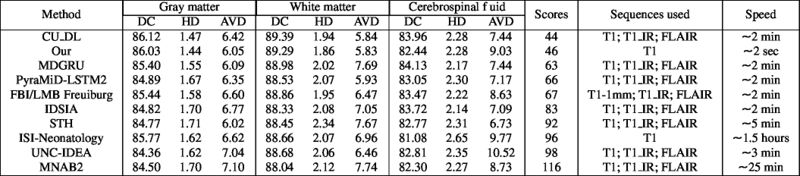
Quantitative results of LOSO experiments in terms of Dice coefficient as compared to the state-of-the-art results. The last one is from P. Moeskops et al. on the 15 test images in MRBrainS13 dataset.
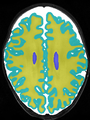
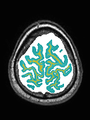
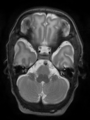
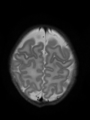
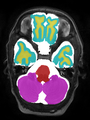
- Some results on axial images at 40 weeks in NeoBrainS12 dataset
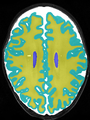
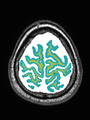
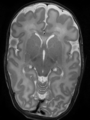
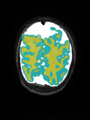
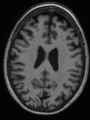
- Some results on coronal images at 30 weeks in NeoBrainS12 dataset
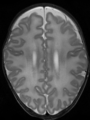
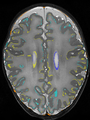
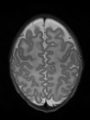
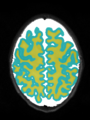
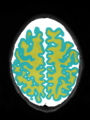
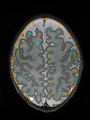
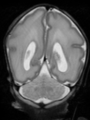
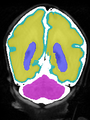
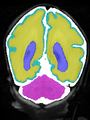
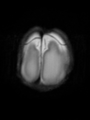
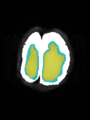
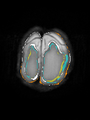
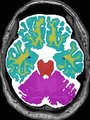
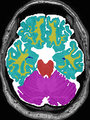
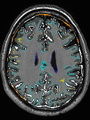
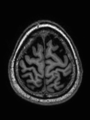
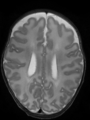
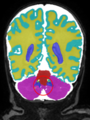
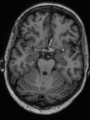
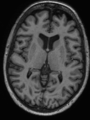
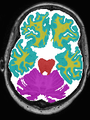
- Some results on axial images of aging adults at 70 ages in MRBrainS13 dataset
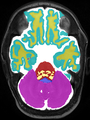
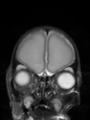
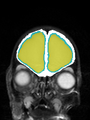
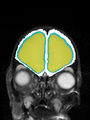
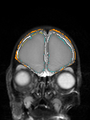
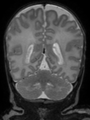
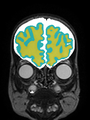
Neonatal brain MR image segmentation
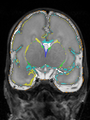
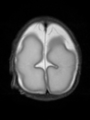
- Results on axial images at 40 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images
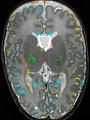
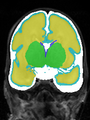
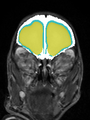
- Results on coronal images at 30 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images (some small errors inside the red circle)
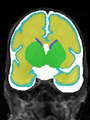
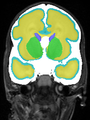
- Results on coronal images at 40 weeks in NeoBrainS12 dataset. More details can be found Here
Some result images (some small errors inside red circles)
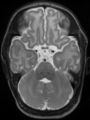
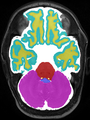
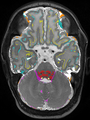
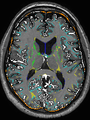
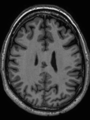
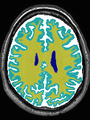
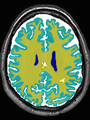
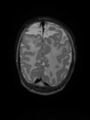
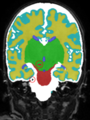
Adult brain MR image segmentation
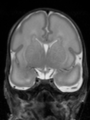
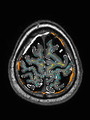
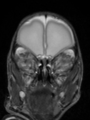
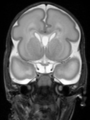
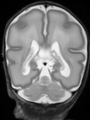
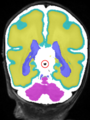
- Results on aging adult images at 70 years in MRBrainS13 dataset. Only top 10 methods among 38 submitted ones are shown. More results and details can be found Here
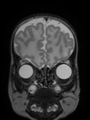
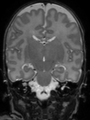
Some result images
Bibtex (lrde.bib)
@InProceedings{ xu.17.icip,
author = {Yongchao Xu and Thierry G\'eraud and Isabelle Bloch},
title = {From Neonatal to Adult Brain {MR} Image Segmentation in a
Few Seconds Using {3D}-Like Fully Convolutional Network and
Transfer Learning},
booktitle = {Proceedings of the 23rd IEEE International Conference on
Image Processing (ICIP)},
year = {2017},
pages = {4417--4421},
month = sep,
address = {Beijing, China},
doi = {10.1109/ICIP.2017.8297117},
abstract = {Brain magnetic resonance imaging (MRI) is widely used to
assess brain developments in neonates and to diagnose a
wide range of neurological diseases in adults. Such studies
are usually based on quantitative analysis of different
brain tissues, so it is essential to be able to classify
them accurately. In this paper, we propose a fast automatic
method that segments 3D brain MR images into different
tissues using fully convolutional network (FCN) and
transfer learning. As compared to existing deep
learning-based approaches that rely either on 2D patches or
on fully 3D FCN, our method is way much faster: it only
takes a few seconds, and only a single modality (T1 or T2)
is required. In order to take the 3D information into
account, all 3 successive 2D slices are stacked to form a
set of 2D color images, which serve as input for the FCN
pre-trained on ImageNet for natural image classification.
To the best of our knowledge, this is the first method that
applies transfer learning to segment both neonatal and
adult brain 3D MR images. Our experiments on two public
datasets show that our method achieves state-of-the-art
results.}
}